41 Biotechnology
On this page
Biotechnology is the use of biological agents for technological advancement. Biotechnology was used for breeding livestock and crops long before people understood the scientific basis of these techniques. Since the discovery of the structure of DNA in 1953, the biotechnology field has grown rapidly through both academic research and private companies. The primary applications of this technology are in medicine (vaccine and antibiotic production) and agriculture (crop genetic modification in order to increase yields). Biotechnology also has many industrial applications, such as fermentation, treating oil spills, and producing biofuels.
DNA and RNA extraction and detection
DNA has two complementary strands linked by hydrogen bonds between the paired bases. Exposure to high temperatures (DNA denaturation) can separate the two strands and cooling can re-anneal them. The DNA polymerase enzyme can replicate the DNA. Unlike DNA, which is located in the eukaryotic cells’ nucleus, RNA molecules leave the nucleus. The most common type of RNA that researchers analyse is the messenger RNA (mRNA) because it represents the protein-coding genes that are actively expressed. However, RNA molecules present some other challenges to analysis, as they are often less stable than DNA.
To study or manipulate nucleic acids, DNA or RNA must be extracted from the cells. Researchers use various techniques to extract different types of DNA (Figure 8.18). Most nucleic acid extraction techniques involve steps to break open the cell and use enzymatic reactions to destroy all macromolecules that are not desired. A lysis buffer (a solution which is mostly a detergent) breaks or lyses the cells. Enzymes such as proteases and ribonucleases (RNAses) break down proteins and RNA, respectively. Using alcohol precipitates the DNA. DNA samples can be frozen at –80°C for years. Scientists perform RNA analysis to study gene expression patterns in cells. RNA is naturally very unstable because RNAses are commonly present in nature and very difficult to inactivate.
Polymerase chain reaction (PCR)
Although genomic DNA is visible to the naked eye when it is extracted in bulk, DNA analysis often requires focusing on one or more specific genome regions. Polymerase chain reaction (PCR) is a technique that scientists use to amplify specific DNA regions for further analysis (Figure 8.19). Researchers use PCR for many purposes in laboratories, such as cloning gene fragments to analyse genetic diseases, identifying contaminant foreign DNA in a sample, and amplifying DNA for sequencing. More practical applications include determining paternity and detecting genetic diseases.
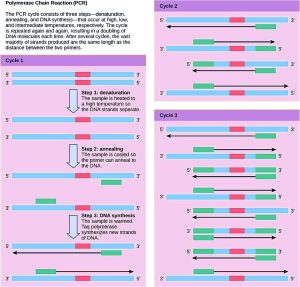
DNA fragments can also be amplified from an RNA template in a process called reverse transcriptase PCR (RT-PCR). The first step is to recreate the original DNA template strand (called cDNA) by applying DNA nucleotides to the mRNA. This process is called reverse transcription. This requires the presence of an enzyme called reverse transcriptase. After the cDNA is made, regular PCR can be used to amplify it.
Dive deeper
Watch this video on PCR: DNA Learning Center. (2010, March 23). Polymerase Chain Reaction (PCR) [YouTube, 1:27mins]
Gel electrophoresis
Because nucleic acids are negatively charged ions at neutral or basic pH in an aqueous environment, an electric field can mobilise them. Gel electrophoresis is a technique that scientists use to separate molecules on the basis of size, using this charge (Figure 8.20). Nucleic acids can be separated as chromosomes or fragments. The nucleic acids are loaded into porous gel matrix and pulled toward the positive electrode at the opposite end. Smaller molecules move through the pores faster than larger molecules. This difference in the migration rate separates the fragments on the basis of size. There are molecular weight standard samples that researchers can run alongside the molecules to provide a size comparison. Nucleic acids in a gel matrix are visible using various fluorescent or coloured dyes. Distinct nucleic acid fragments appear as bands at specific distances from the top. A mixture of genomic DNA fragments of varying sizes appear as a long smear; whereas, uncut genomic DNA is usually too large to run through the gel and forms a single large band at the top.
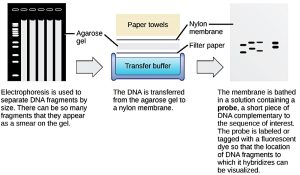
Nucleic acids in samples can be probed for the presence of certain sequences by designing and labelling short DNA fragments, or probes with radioactive or fluorescent dyes. Gel electrophoresis separates the nucleic acid fragments according to their size. Transferring fragments from a gel to a nylon membrane is known as blotting (Figure 8.19). Scientists can then probe the nucleic acid fragments that are bound to the membrane’s surface with specific radioactively or fluorescent labeled probe sequences. When scientists transfer DNA to a nylon membrane, they refer to the technique as Southern blotting. When they transfer the RNA to a nylon membrane, they call it Northern blotting. Scientists use Southern blots to detect the presence of certain DNA sequences in a given genome, and Northern blots to detect gene expression.
Molecular cloning
In general, the word cloning means the creation of a perfect replica; however, in biology, the re-creation of a whole organism is referred to as reproductive cloning. Similarly, molecular cloning is to reproduce desired regions or fragments of the genome.
 Case study
Case study
A new technique for producing synthetic insulin (previously produced in pig and cow pancreases) provided hope for diabetes treatment, addressing the shortage of insulin derived from animals. This breakthrough spurred the “BioTech Boom,” leading to increased research and funding for innovative health solutions.
Cloning small genome fragments allows researchers to manipulate and study specific genes (and their protein products), or noncoding regions in isolation.
A plasmid, or vector, is a small circular DNA molecule that replicates independently of the chromosomal DNA.
In cloning, plasmid molecules are used to insert a desired DNA fragment. Plasmids are usually introduced into a bacterial host for proliferation. The DNA fragment from the studied organism is known as a transgene.
Plasmids occur naturally in bacterial populations (such as Escherichia coli) and have genes that can contribute favourable traits to the organism, such as antibiotic resistance (the ability to be unaffected by antibiotics). Plasmids have repurposed and engineered as vectors for molecular cloning and the large-scale production of important reagents, such as insulin and human growth hormone.
Restriction endonucleases recognise specific DNA sequences and cut them in a predictable manner. They are naturally produced by bacteria as a defense mechanism against foreign DNA. Many restriction endonucleases make staggered cuts in the two DNA strands, such that the cut ends have a 2- or 4-base single-stranded overhang known as sticky ends. Adding the enzyme DNA ligase permanently joins the DNA fragments via phosphodiester bonds. In this way, we can splice any DNA fragment generated by restriction endonuclease cleavage between the plasmid DNA’s two ends that has been cut with the same restriction endonuclease (Figure 8.21).
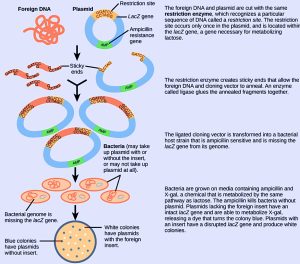
Plasmids with foreign DNA inserted into them are called recombinant DNA molecules because they are created artificially and do not occur in nature. Recombinant proteins are proteins that are expressed from recombinant DNA molecules. Not all recombinant plasmids are capable of expressing genes. The recombinant DNA may need to move into a different vector (or host) that is better designed for gene expression. Scientists may also engineer plasmids to express proteins only when certain environmental factors stimulate them, so they can control the recombinant proteins’ expression.
 Case study
Case study
Enviro-Pigs
EnviroPigs are genetically modified Yorkshire pigs developed to reduce environmental phosphorus pollution. Scientists introduced a gene from E. coli and a mouse into the pig’s genome, enabling the production of phytase in their saliva. This enzyme breaks down phytate, the main form of phosphorus in grains, making it digestible. As a result, EnviroPigs excrete significantly less phosphorus, reducing the environmental impact of pig farming, particularly eutrophication in water bodies. This innovation represents a sustainable approach to livestock production, although regulatory and public acceptance challenges have limited their commercial deployment.
Biosteel Goats
Biosteel goats are transgenic animals engineered to produce spider silk proteins in their milk. By inserting genes from orb-weaving spiders into goats, researchers enabled the production of silk proteins that can be harvested and spun into fibers. These fibers, known as Biosteel, are incredibly strong, lightweight, and biodegradable, with potential applications in medicine (e.g., sutures), defense (e.g., body armor), and industry (e.g., ropes and cables). The use of goats as bioreactors offers a scalable method for producing spider silk, which is otherwise difficult to harvest in large quantities from spiders themselves.
Transgenic Mice
Transgenic mice are invaluable tools in biomedical research, particularly for studying cancer and genetic diseases. By introducing or knocking out specific genes, such as oncogenes or tumour suppressors like p53, researchers can model human diseases in a controlled environment. Mice with p53 mutations, for example, develop spontaneous tumours, providing insights into cancer development, progression, and treatment responses. These models allow scientists to test new drugs, study gene function, and understand disease mechanisms at the molecular level. Their genetic similarity to humans and short reproductive cycles make them ideal for translational research.
Transgenic Cows
Transgenic cows engineered to express modified kappa-casein genes produce milk with improved cheese-making properties. Kappa-casein plays a crucial role in milk coagulation, and its enhanced expression leads to better curd formation, higher cheese yield, and improved processing efficiency. These cows are developed using gene-editing techniques to insert bovine kappa-casein variants associated with superior milk quality. This innovation benefits the dairy industry by increasing productivity and reducing waste. Additionally, such modifications can be tailored to produce milk with specific nutritional or hypoallergenic properties, offering potential health benefits to consumers.
GloFish as Biosensors
GloFish are genetically modified fluorescent zebrafish originally developed as environmental biosensors. By inserting genes from jellyfish or coral that produce fluorescent proteins, scientists created fish that glow under UV light. Some GloFish lines are designed to fluoresce in response to environmental toxins, such as heavy metals or pollutants, making them useful for detecting water contamination. Although now popular as ornamental pets, their original purpose was to serve as living indicators of ecosystem health. GloFish demonstrate how genetic engineering can be applied to environmental monitoring, combining biological sensitivity with visual detection.

